PCA Explained
In Machine Learning and Statistics, Principal Component Analysis (PCA or linear PCA) is a classic and widely used technique for data transformation. PCA can, for example, transform your high dimensional data into points in two or three dimensional space so that they can be easily visualized in a scatter plot, or it could be a preprocessing step in supervised learning tasks (classification, regression) so that the data is projected in low dimensional space where the new features are decorrelated, leading to potentially better accuracy. This tutorial will explain all the math behind PCA and provide an example where you’ll practice step-by-step how to perform PCA on a synthetic data matrix.
Prerequisite
Here it is assumed that you have some basic understanding of Linear Algebra (e.g. matrix arithmetics, the geometric interpretation of inner product), Calculus (e.g. Lagrangian Multiplier) and Statistics (e.g. what is mean and variance). In addition, definitions of some math concepts and theorems (listed below) are needed to prove the properties of the resulting transformed data matrix.
Definition 1
A scalar is called an eigenvalue of a matrix if there is a nontrivial (non-zero) solution of . Such an is called an eigenvector corresponding to the eigenvalue .
Definition 2
A real-valued symmetric matrix is said to be positive semidefinite if the scalar is non-negative for every non-zero real-valued column vector .
Theorem 1
If is a real-valued symmetric matrix, then the eigenvectors corresponding to different eigenvalues must be orthogonal to each other.
[http://www.math.hawaii.edu/~lee/linear/eigen.pdf]
Theorem 2
A real-valued symmetric matrix is positive semidefinite if and only if all of its eigenvalues are non-negative.
[http://theanalysisofdata.com/probability/C4.html]
The Math
Let’s say we have a data matrix where is the number of observations and is the number of features — each observation is described by attributes.
Now we wish to describe each observation by a single attribute. In other words, the matrix will be turned into a column matrix (i.e. vector), and we want the variance of the sample composed of the elements from this vector to be as large as possible.
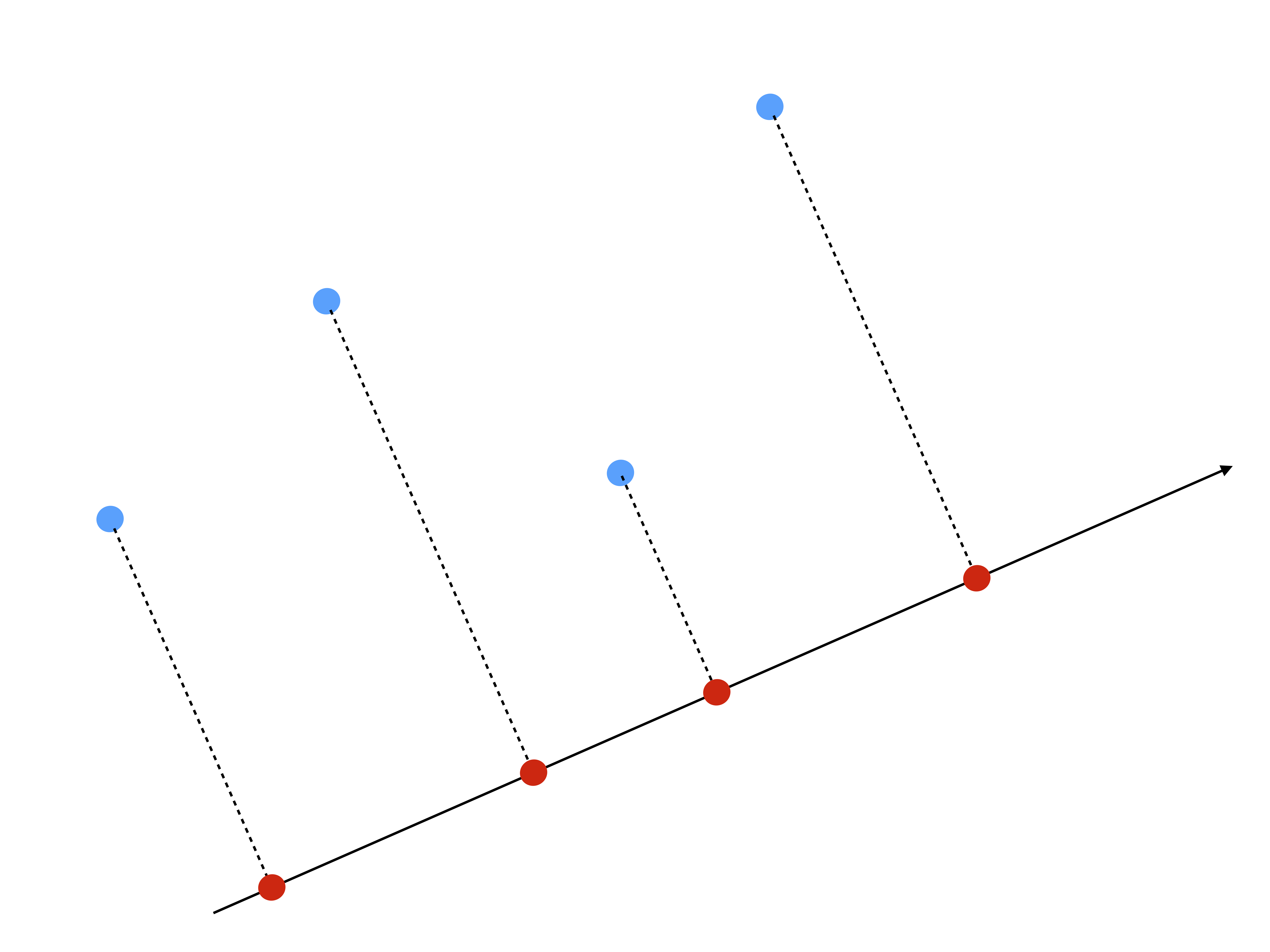
Project points from 2-D space to 1-D space
One way to do this (in the case of PCA) is to project the collection of points in -dimensional space into 1-dimensional space, or equivalently right-multiply with a column vector
giving rise to the transformed data matrix
Note: For mathmatical convenience, the data matrix is assumed to have been centered (each column sums to one): , where
In case had not been centered, you can simply subtract the column-mean from each element in each column.
Given that has been centered, the transformed data matrix will be centered as well:
Remember we wanted to make the variance of the sample composed of the elements from to be as large as possible. In other words, we want to maximize (over the space of all ), which can be expressed in terms of and :
Here we can see that is proportional to the norm of , which in turn depends on the norm of . If we were to allow the norm of to be unbounded, the could be arbitrarily large!
To fix this we need to put some constraint on — we make it a unit-length vector:
So this turns the original function optimization problem into one with an equality constraint, which can be solved using Lagrangian Multiplier:
Taking the derivative of the Lagrangian function with respect to and set it to zero,
we end up with the condition that must statisfy:
where
According to Defitition 1, must be an eigenvector of corresponding to eigenvalue
We can further show that the variance of happens to be equal to :
The matrix (i.e. the covariance matrix of ) has the following properties:
-
is symmetric:
-
is semi-positive definite: For any real-valued non-zero vector ,
Suppose are the eigenvalues of , and are the sets of eigenvectors corresponding to each eigenvalue. Then must be one of the eigenvalues, and must be an eigenvector corresponding to .
According to Theorem 1, we know that eigenvectors are orthogonal to each other.
According to Theorem 2, we know that the eigenvalues are non-negative —
Based on the properties of , , , and , we can apply PCA on in the following steps:
- Compute the covariance matrix , where should have been centered.
- Find the eigenvalues of , as well as the corresponding eigenvectors . The eigenvalues should be in descending order:
- Determine the number of features to keep in the transformed matrix ().
- Build the matrix
- Compute the transformed matrix .
And we can prove that the columns of have the properties that we desired:
- has the largest variance , followed by with variance , and so on.
- The columns of are orthogonal with each other — Consider and where ). It follows that
Example
We conclude our discussion with an example where we’ll generate a synthetic dataset and apply PCA as prescribed above.
First we will generate 2-D dataset with covariance matrix and mean (shown in the figure below):
import numpy as np
import matplotlib.pyplot as plt
def generate_2Ddata(size, covariance, theta, offset):
X = np.random.multivariate_normal([0, 0], covariance, size)
rot_mat = np.array([[np.cos(theta), -np.sin(theta)],
[np.sin(theta), np.cos(theta)]])
return X.dot(rot_mat) + offset
size = 1000
X = generate_2Ddata(size, [[0.1, 0.], [0., 1.]], np.pi/4, [-1., 1.])
Compute covariance matrix of a centered data matrix
# Center the data
X = X - X.mean(axis=0)
# Compute covariance matrix
sigma = X.T.dot(X) / (size - 1)
Find the eigenvalues and eigenvectors, and sort them in descending order of eigenvalues.
# eigval[i] is the ith eigenvalue
# eigvec[:, i] is the eigenvector corresponding to eigval[i]
eigval, eigvec = np.linalg.eig(sigma)
indices = np.argsort(-eigval)
eigvec = eigvec[:, indices]
For now we’ll keep all principle components, but you can keep the top k components with largest variance:
eigvec = eigvec[:, :k]
Apply PCA as a linear transformation on X
X_pca = X.dot(eigvec)
Display PCA-transfomred data matrix in scatter plot
plt.scatter(X_pca[:, 0], X_pca[:, 1])
plt.xlim([-5, 5])
plt.ylim([-5, 5])
plt.xlabel('1st Principal Component')
plt.ylabel('2nd Principal Component')
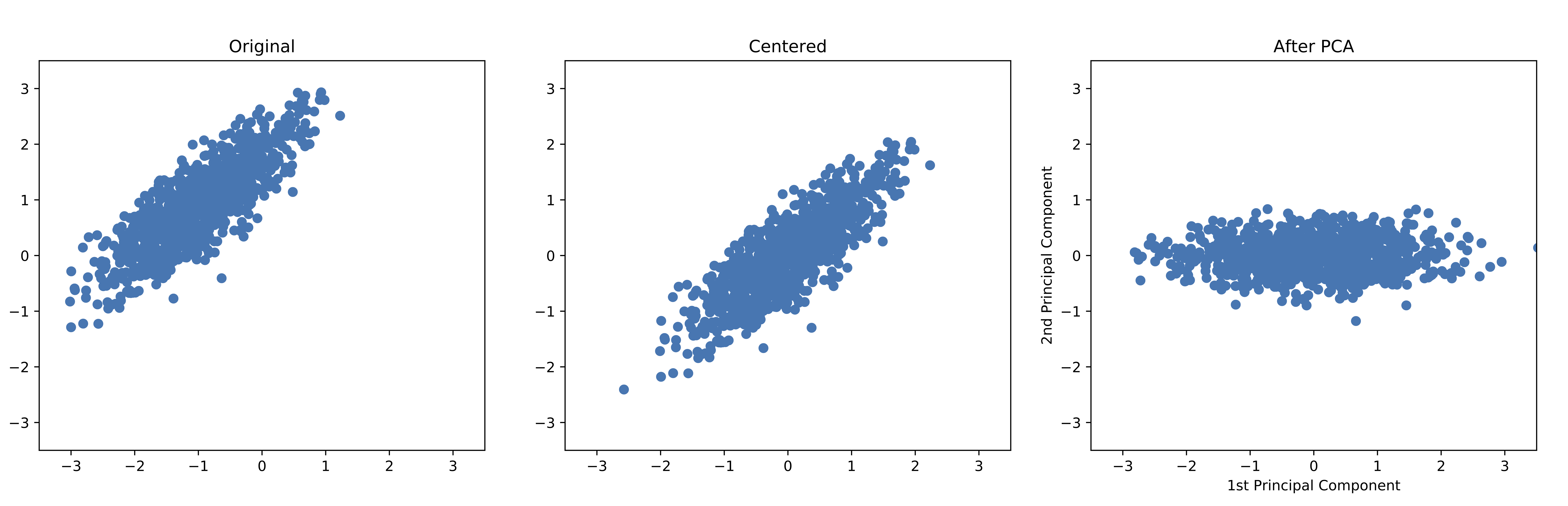
Left: Original, uncentered. Middle: Centered. Right: PCA-transformed